Linear regression with a two-level factor explanatory variable
Last updated on 2024-03-12 | Edit this page
Overview
Questions
- How can we explore the relationship between one continuous variable and one categorical variable with two groups prior to fitting a simple linear regression?
- How can we fit a simple linear regression model with one two-level categorical explanatory variable in R?
- How does the use of the simple linear regression equation differ between the continuous and categorical explanatory variable cases?
- How can the parameters of this model be interpreted in R?
- How can this model be visualised in R?
Objectives
- Use the ggplot2 package to explore the relationship between a continuous variable and a two-level factor variable.
- Use the lm command to fit a simple linear regression with a two-level factor explanatory variable.
- Distinguish between the baseline and contrast levels of the categorical variable.
- Use the jtools package to interpret the model output.
- Use the jtools and ggplot2 packages to visualise the resulting model.
In this episode we will study linear regression with one two-level
categorical variable. We can explore the relationship between two
variables ahead of fitting a model using the ggplot2
package.
Exploring the relationship between a continuous variable and a two-level categorical variable
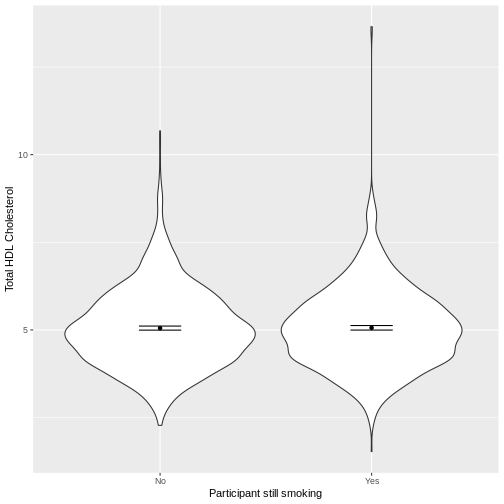
Let us take SmokeNow and TotChol as an
example. SmokeNow describes whether someone who has smoked
> 100 cigarettes in their life is currently smoking.
TotChol describes the total HDL cholesterol in someone’s
blood. In the code below, we first remove rows with missing values using
drop_na() from the tidyr package. We then
initiate a plotting object using ggplot. We select the
variables of interest inside aes(). We then make a violin
plot using geom_violin. The shapes of the objects are
representative of the distributions of TotChol in the two
groups. We overlay the means and their 95% confidence intervals using
stat_summary(). Finally, we change the axis labels using
xlab() and ylab().
R
dat %>%
drop_na(c(SmokeNow, TotChol)) %>%
ggplot(aes(x = SmokeNow, y = TotChol)) +
geom_violin() +
stat_summary(fun = "mean", size = 0.2) +
stat_summary(fun.data = "mean_cl_normal", geom = "errorbar", width = 0.2) +
xlab("Participant still smoking") +
ylab("Total HDL Cholesterol")
Notes on thefunandfun.dataarguments instat_summary()
The fun and fun.data arguments both apply
statistical operations to data but do slightly different things.
fun takes data as vectors and will return single values for
each these vectors. In the above example, we calculate the mean for each
vector (each SmokeNow group). fun.data expects
a dataset (which may be a simple vector) and provides three values for
each dataset: y, ymin and ymax.
In our case, ymin is lower bound of the confidence interval and ymax is
the upper bound of the confidence interval.
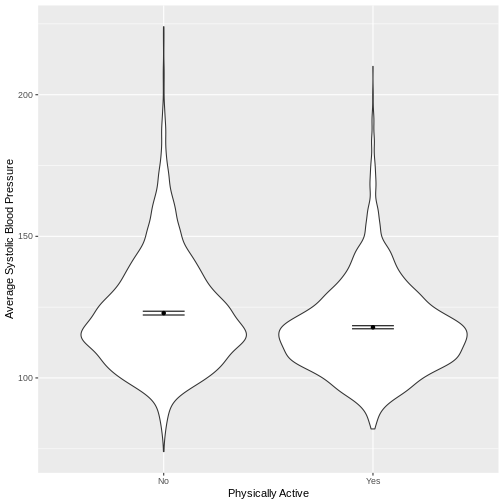
Exercise
You have been asked to model the relationship between average systolic blood pressure and physical activity in the NHANES data. Use the ggplot2 package to create an exploratory plot, ensuring that it includes the following elements:
- Average systolic blood pressure (
BPSysAve) on the y-axis and physical activity (PhysActive) on the x-axis, from the NHANES data. - These data presented using a violin plot.
- The y-axis labelled as “Average Systolic Blood Pressure” and the x-axis labelled as “Physically Active”.
R
dat %>%
drop_na(c(PhysActive, BPSysAve)) %>%
ggplot(aes(x = PhysActive, y = BPSysAve)) +
geom_violin() +
stat_summary(fun = "mean", size = 0.2) +
stat_summary(fun.data = "mean_cl_normal", geom = "errorbar", width = 0.2) +
xlab("Physically Active") +
ylab("Average Systolic Blood Pressure")
Fitting and interpreting a simple linear regression model with one two-level categorical variable
We proceed to fit a linear regression model using the
lm() command, as we did in the previous episode. The model
is then interpreted using summ().
Note that even though we are using the same equation of the simple
linear regression model (\(E(y) = \beta_0 +
\beta_1 \times x_1\)), our interpretation of the
summ() output differs slightly from our interpretation in
the previous episode. Recall that our categorical explanatory variable
has two levels, "No" and "Yes". One of these
is taken by the model as the baseline ("No"), the other as
the contrast ("Yes"). The first level alphabetically is
chosen by R as the baseline, unless specified otherwise.
The intercept in the summ() output is the estimated mean
for the baseline, i.e. for participants that stopped smoking. The
SmokeNowYes estimate is the estimated average difference in
TotChol between participants that stopped smoking and
participants that did not stop smoking. We can therefore write the
equation for this model as:
\[E(\\text{Total HDL cholesterol}) = 5.053 + 0.008 \\times x\_1\],
where \(x_1 = 1\) if a participant has continued to smoke and 0 otherwise (i.e. the participant stopped smoking).
R
TotChol_SmokeNow_lm <- dat %>%
lm(formula = TotChol ~ SmokeNow)
summ(TotChol_SmokeNow_lm, confint = TRUE, digits = 3)
OUTPUT
MODEL INFO:
Observations: 2741 (7259 missing obs. deleted)
Dependent Variable: TotChol
Type: OLS linear regression
MODEL FIT:
F(1,2739) = 0.031, p = 0.861
R² = 0.000
Adj. R² = -0.000
Standard errors: OLS
------------------------------------------------------------
Est. 2.5% 97.5% t val. p
----------------- ------- -------- ------- --------- -------
(Intercept) 5.053 4.995 5.111 170.707 0.000
SmokeNowYes 0.008 -0.078 0.094 0.175 0.861
------------------------------------------------------------Exercise
- Using the
lm()command, fit a simple linear regression of average systolic blood pressure (BPSysAve)
as a function of physical activity (PhysActive). Name thislmobjectBPSysAve_PhysActive_lm. - Using the
summ()function from thejtoolspackage, answer the following questions:
- What average systolic blood pressure does the model predict, on
average, for an individual who is not physically active?
- By how much is average systolic blood pressure expected to change,
on average, for a physically active individual?
- Given these two values and the names of the response and explanatory variables, how can the general equation \(E(y) = \beta_0 + {\beta}_1 \times x_1\) be adapted to represent this model?
R
BPSysAve_PhysActive_lm <- dat %>%
lm(formula = BPSysAve ~ PhysActive)
summ(BPSysAve_PhysActive_lm, confint = TRUE, digits = 3)
OUTPUT
MODEL INFO:
Observations: 6781 (3219 missing obs. deleted)
Dependent Variable: BPSysAve
Type: OLS linear regression
MODEL FIT:
F(1,6779) = 133.611, p = 0.000
R² = 0.019
Adj. R² = 0.019
Standard errors: OLS
-------------------------------------------------------------------
Est. 2.5% 97.5% t val. p
------------------- --------- --------- --------- --------- -------
(Intercept) 122.892 122.275 123.509 390.285 0.000
PhysActiveYes -5.002 -5.851 -4.154 -11.559 0.000
-------------------------------------------------------------------- 122.892 mmHg
- Decrease by 5.002 mmHg
- \(E(\text{BPSysAve}) = 122.892 - 5.002 \times x_1\), where \(x_1 = 0\) if an individual is not physically active and \(x_1 = 1\) if an individual is physically active.
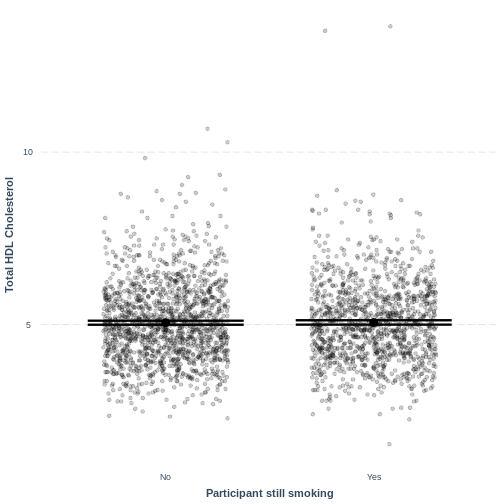
Visualising a simple linear regression model with one two-level categorical explanatory variable
Finally, we visually inspect the parameter estimates provided by our
model. Again we can use effect_plot() from the
jtools package. We include jitter = c(0.3, 0)
and point.alpha = 0.2 so that points are spread out
HORIZONTALLY and so that multiple overlayed points create a darker
colour, respectively. The resulting plot differs from the scatterplot
obtained in the previous episode. Here, the plot shows the mean Total
HDL Cholesterol estimates for each level of SmokeNow, with
their 95% confidence intervals. This allows us to see how different the
means are predicted to be and within what range we can expect the true
population means to fall.
R
effect_plot(TotChol_SmokeNow_lm, pred = SmokeNow,
plot.points = TRUE, jitter = c(0.3, 0), point.alpha = 0.2) +
xlab("Participant still smoking") +
ylab("Total HDL Cholesterol")
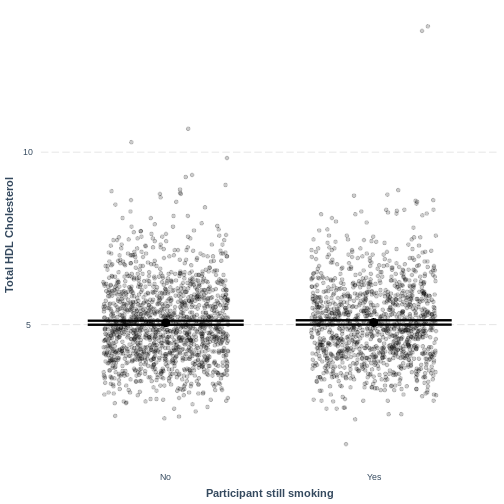
Notes onjitterandpoint.alpha
Including jitter = c(0.3, 0) results in points being
randomly jittered horizontally. Therefore, your plot will
differ slightly from the one shown above. Re-running the code will also
give a slightly different jitter. If you would want to fix the
jitter to one randomisation, you could run a
set.seed() command ahead of effect_plot.
set.seed() takes one positive value, which specifies the
randomisation. This can be any positive value. For example, if you run
the code below, your jitter will match the one shown on this page:
R
set.seed(20) #fix the jitter to a particular pattern
effect_plot(TotChol_SmokeNow_lm, pred = SmokeNow,
plot.points = TRUE, jitter = c(0.3, 0), point.alpha = 0.2) +
xlab("Participant still smoking") +
ylab("Total HDL Cholesterol")
Including point.alpha = 0.2 introduces opacity into the
plotted points. As a result, if many points are plotted on top of each
other, this area shows up with darker dots. Including opacity allows us
to see where many of the points lie, which is handy with big public
health data sets.
R
effect_plot(BPSysAve_PhysActive_lm, pred = PhysActive,
plot.points = TRUE, jitter = c(0.3, 0), point.alpha = 0.2) +
xlab("Physically Active") +
ylab("Average Systolic Blood Pressure")
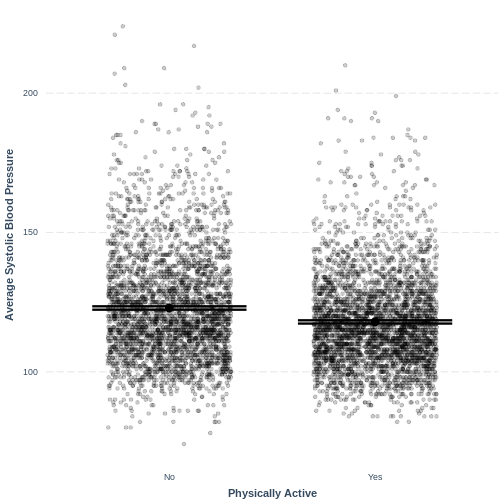
This plot shows the mean estimates for BPSysAve for the
two groups, alongside their 95% confidence intervals. The mean estimates
are represented by the Intercept for the non-physically
active group and by Intercept + PhysActiveYes
for the physically active group.
Key Points
- As a first exploration of the data, construct a violin plot describing the relationship between the two variables.
- Use
lm()to fit a simple linear regression model. - Use
summ()to obtain parameter estimates for the model. - The intercept estimates the mean in the outcome variable for the baseline group. The other parameter estimates the difference in the means in the outcome variable between the baseline and contrast group.
- Use
effect_plot()to visualise the estimated means per group along with their 95% CIs.
