Making Plots With plotnine
Last updated on 2023-05-08 | Edit this page
Estimated time: 90 minutes
Overview
Questions
- How can I visualize data in Python?
- What is ‘grammar of graphics’?
Objectives
- Create a
plotnineobject. - Set universal plot settings.
- Modify an existing plotnine object.
- Change the aesthetics of a plot such as color.
- Edit the axis labels.
- Build complex plots using a step-by-step approach.
- Create scatter plots, box plots, and time series plots.
- Use the facet_wrap and facet_grid commands to create a collection of plots splitting the data by a factor variable.
- Create customized plot styles to meet their needs.
Disclaimer
Python has powerful built-in plotting capabilities such as
matplotlib, but for this episode, we will be using the plotnine package,
which facilitates the creation of highly-informative plots of structured
data based on the R implementation of ggplot2 and The
Grammar of Graphics by Leland Wilkinson. The plotnine
package is built on top of Matplotlib and interacts well with
Pandas.
Reminder
plotnine is not included in the standard Anaconda
installation and needs to be installed separately. If you haven’t done
so already, you can find installation instructions on the Setup
page.
Just as with the other packages, plotnine needs to be
imported. It is good practice to not just load an entire package such as
from plotnine import *, but to use an abbreviation as we
used pd for Pandas:
From now on, the functions of plotnine are available
using p9.. For the exercise, we will use the
surveys.csv data set, with the NA values
removed
Plotting with plotnine
The plotnine package (cfr. other packages conform The
Grammar of Graphics) supports the creation of complex plots from data in
a dataframe. It uses default settings, which help creating publication
quality plots with a minimal amount of settings and tweaking.
plotnine graphics are built step by step by adding new
elements adding different elements on top of each other using the
+ operator. Putting the individual steps together in
brackets () provides Python-compatible syntax.
To build a plotnine graphic we need to:
- Bind the plot to a specific data frame using the
dataargument:
As we have not defined anything else, just an empty figure is available and presented.
- Define aesthetics (
aes), by selecting variables used in the plot andmappingthem to a presentation such as plotting size, shape, color, etc. You can interpret this as: which of the variables will influence the plotted objects/geometries:
The most important aes mappings are: x, y,
alpha, color, colour,
fill, linetype, shape,
size and stroke.
- Still no specific data is plotted, as we have to define what kind of
geometry will be used for the plot. The most straightforward is probably
using points. Points is one of the
geomsoptions, the graphical representation of the data in the plot. Others are lines, bars,… To add a geom to the plot use+operator:
PYTHON
(p9.ggplot(data=surveys_complete,
mapping=p9.aes(x='weight', y='hindfoot_length'))
+ p9.geom_point()
)The + in the plotnine package is
particularly useful because it allows you to modify existing
plotnine objects. This means you can easily set up plot
templates and conveniently explore different types of plots, so
the above plot can also be generated with code like this:
PYTHON
# Create
surveys_plot = p9.ggplot(data=surveys_complete,
mapping=p9.aes(x='weight', y='hindfoot_length'))
# Draw the plot
surveys_plot + p9.geom_point()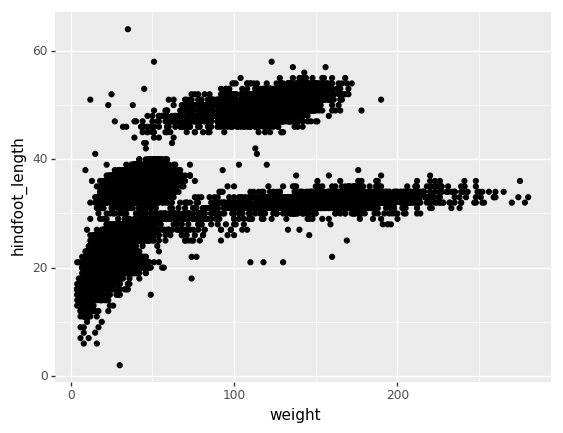
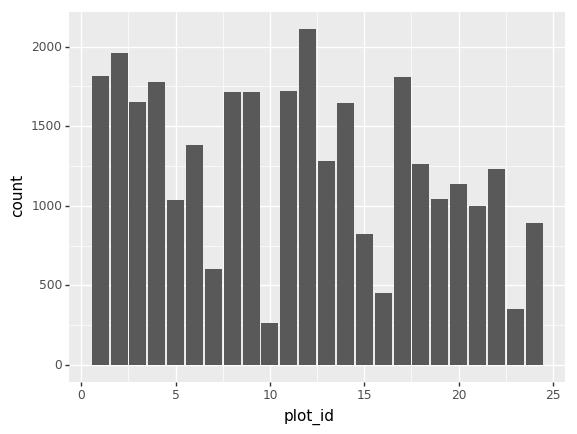
Challenge - bar chart
Working on the surveys_complete data set, use the
plot-id column to create a bar-plot that
counts the number of records for each plot. (Check the documentation of
the bar geometry to handle the counts)
Notes:
- Anything you put in the
ggplot()function can be seen by any geom layers that you add (i.e., these are universal plot settings). This includes thexandyaxis you set up inaes(). - You can also specify aesthetics for a given
geomindependently of the aesthetics defined globally in theggplot()function.
Building your plots iteratively
Building plots with plotnine is typically an iterative
process. We start by defining the dataset we’ll use, lay the axes, and
choose a geom. Hence, the data, aes and
geom-* are the elementary elements of any graph:
PYTHON
(p9.ggplot(data=surveys_complete,
mapping=p9.aes(x='weight', y='hindfoot_length'))
+ p9.geom_point()
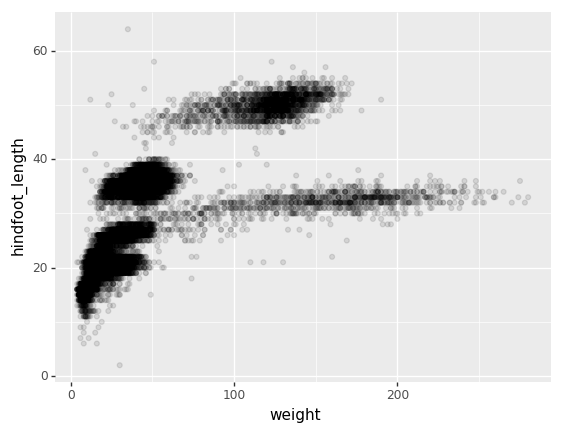
)Then, we start modifying this plot to extract more information from it. For instance, we can add transparency (alpha) to avoid overplotting:
PYTHON
(p9.ggplot(data=surveys_complete,
mapping=p9.aes(x='weight', y='hindfoot_length'))
+ p9.geom_point(alpha=0.1)
)
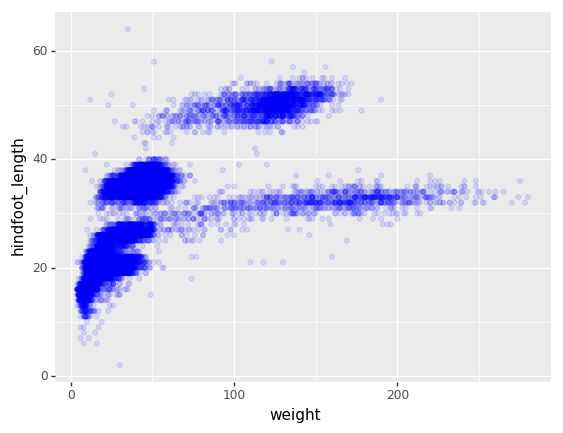
We can also add colors for all the points
PYTHON
(p9.ggplot(data=surveys_complete,
mapping=p9.aes(x='weight', y='hindfoot_length'))
+ p9.geom_point(alpha=0.1, color='blue')
)
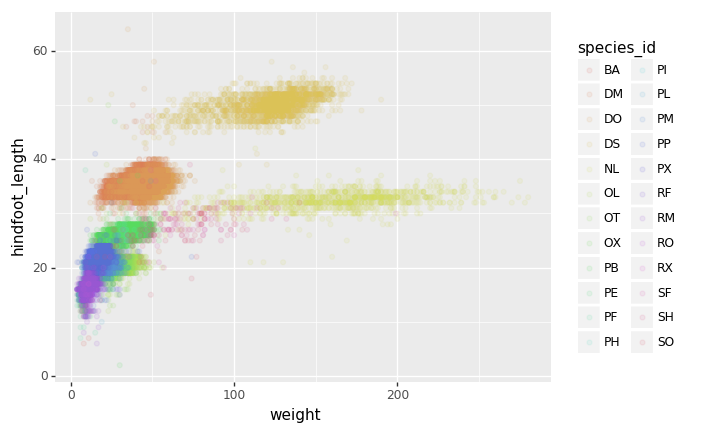
Or to color each species in the plot differently, map the
species_id column to the color aesthetic:
PYTHON
(p9.ggplot(data=surveys_complete,
mapping=p9.aes(x='weight',
y='hindfoot_length',
color='species_id'))
+ p9.geom_point(alpha=0.1)
)
Apart from the adaptations of the arguments and settings of the
data, aes and geom-* elements,
additional elements can be added as well, using the +
operator:
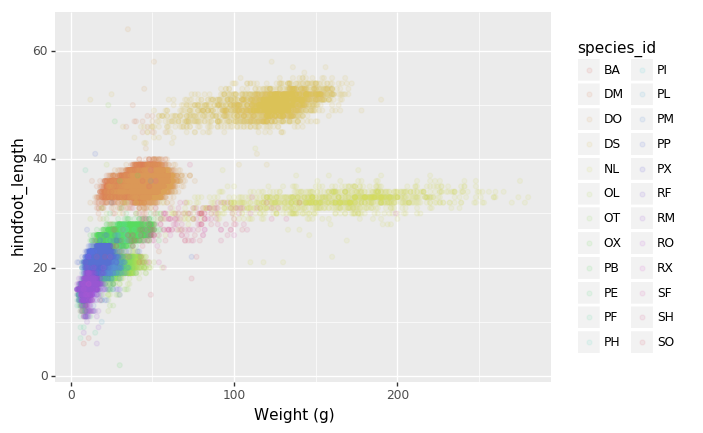
- Changing the labels:
PYTHON
(p9.ggplot(data=surveys_complete,
mapping=p9.aes(x='weight', y='hindfoot_length', color='species_id'))
+ p9.geom_point(alpha=0.1)
+ p9.xlab("Weight (g)")
)
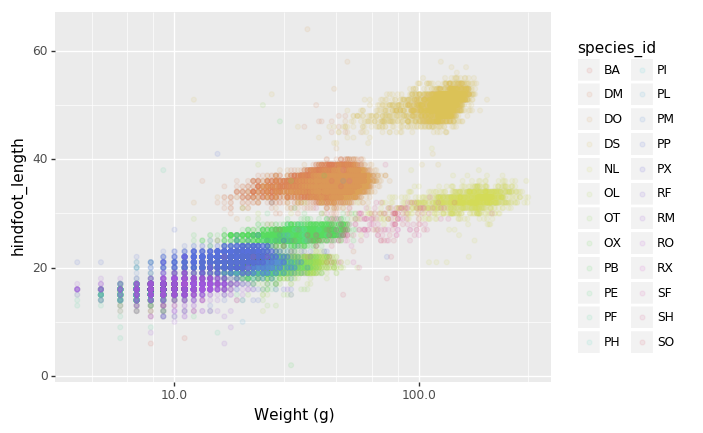
- Defining scale for colors, axes,… For example, a log-version of the x-axis could support the interpretation of the lower numbers:
PYTHON
(p9.ggplot(data=surveys_complete,
mapping=p9.aes(x='weight', y='hindfoot_length', color='species_id'))
+ p9.geom_point(alpha=0.1)
+ p9.xlab("Weight (g)")
+ p9.scale_x_log10()
)
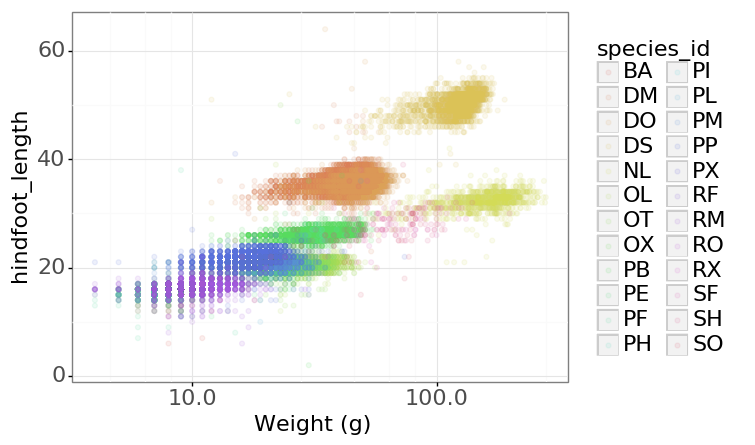
- Changing the theme (
theme_*) or some specific theming (theme) elements. Usually plots with white background look more readable when printed. We can set the background to white using the functiontheme_bw().
PYTHON
(p9.ggplot(data=surveys_complete,
mapping=p9.aes(x='weight', y='hindfoot_length', color='species_id'))
+ p9.geom_point(alpha=0.1)
+ p9.xlab("Weight (g)")
+ p9.scale_x_log10()
+ p9.theme_bw()
+ p9.theme(text=p9.element_text(size=16))
)
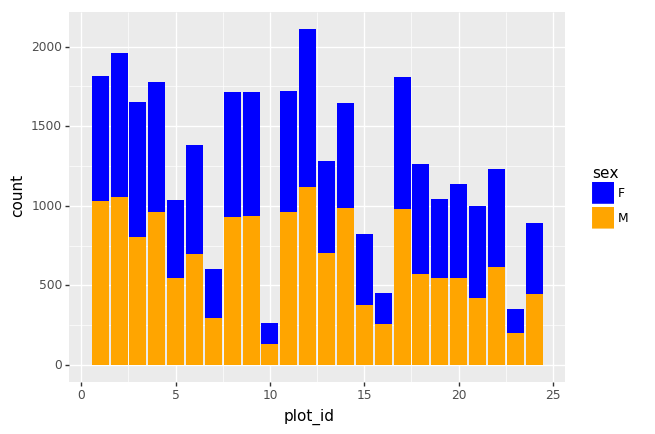
Challenge - Bar plot adaptations
Adapt the bar plot of the previous exercise by mapping the
sex variable to the color fill of the bar chart. Change the
scale of the color fill by providing the colors
blue and orange manually (see API
reference to find the appropriate function).
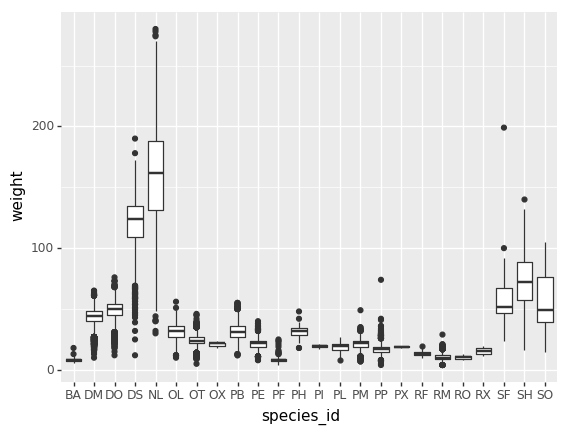
Plotting distributions
Visualizing distributions is a common task during data exploration
and analysis. To visualize the distribution of weight
within each species_id group, a boxplot can be used:
PYTHON
(p9.ggplot(data=surveys_complete,
mapping=p9.aes(x='species_id',
y='weight'))
+ p9.geom_boxplot()
)
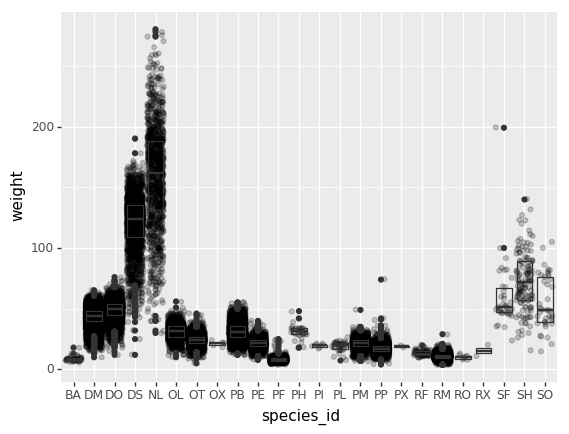
By adding points of the individual observations to the boxplot, we can have a better idea of the number of measurements and of their distribution:
PYTHON
(p9.ggplot(data=surveys_complete,
mapping=p9.aes(x='species_id',
y='weight'))
+ p9.geom_jitter(alpha=0.2)
+ p9.geom_boxplot(alpha=0.)
)
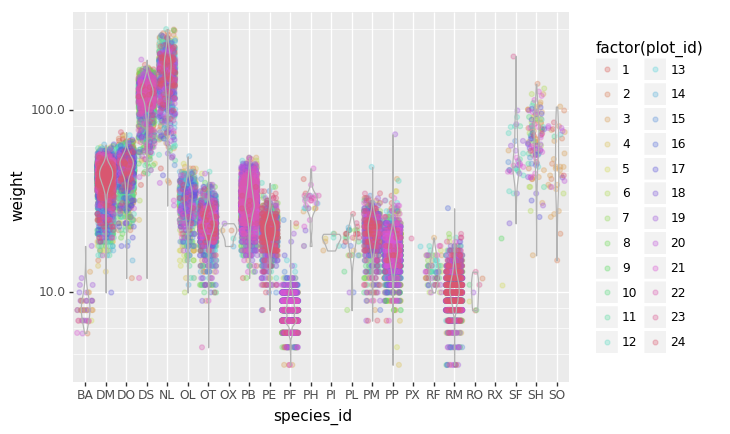
Challenge - distributions
Boxplots are useful summaries, but hide the shape of the distribution. For example, if there is a bimodal distribution, this would not be observed with a boxplot. An alternative to the boxplot is the violin plot (sometimes known as a beanplot), where the shape (of the density of points) is drawn.
In many types of data, it is important to consider the scale of the observations. For example, it may be worth changing the scale of the axis to better distribute the observations in the space of the plot.
- Replace the box plot with a violin plot, see
geom_violin() - Represent weight on the log10 scale, see
scale_y_log10() - Add color to the datapoints on your boxplot according to the plot
from which the sample was taken (
plot_id)
Hint: Check the class for plot_id. By using
factor() within the aes mapping of a variable,
plotnine will handle the values as category values.
Plotting time series data
Let’s calculate number of counts per year for each species. To do
that we need to group data first and count the species
(species_id) within each group.
PYTHON
yearly_counts = surveys_complete.groupby(['year', 'species_id'])['species_id'].count()
yearly_countsWhen checking the result of the previous calculation, we actually
have both the year and the species_id as a row
index. We can reset this index to use both as column variable:
Timelapse data can be visualised as a line plot
(geom_line) with years on x axis and counts on
the y axis.
Unfortunately this does not work, because we plot data for all the
species together. We need to tell plotnine to draw a line
for each species by modifying the aesthetic function and map the
species_id to the color:
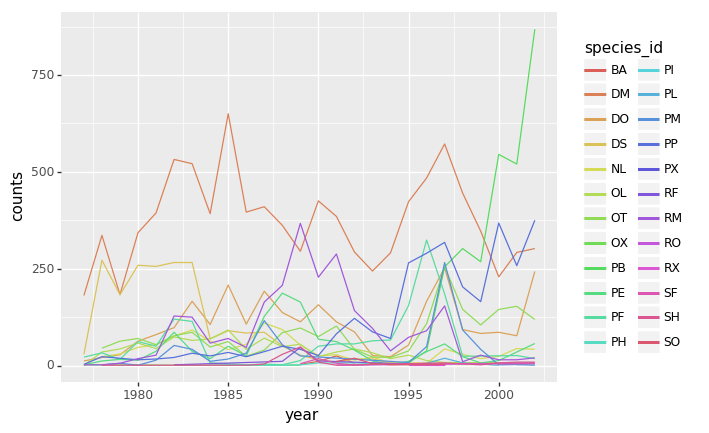
PYTHON
(p9.ggplot(data=yearly_counts,
mapping=p9.aes(x='year',
y='counts',
color='species_id'))
+ p9.geom_line()
)
Faceting
As any other library supporting the Grammar of Graphics,
plotnine has a special technique called faceting
that allows to split one plot into multiple plots based on a factor
variable included in the dataset.
Consider our scatter plot of the weight versus the
hindfoot_length from the previous sections:
PYTHON
(p9.ggplot(data=surveys_complete,
mapping=p9.aes(x='weight',
y='hindfoot_length',
color='species_id'))
+ p9.geom_point(alpha=0.1)
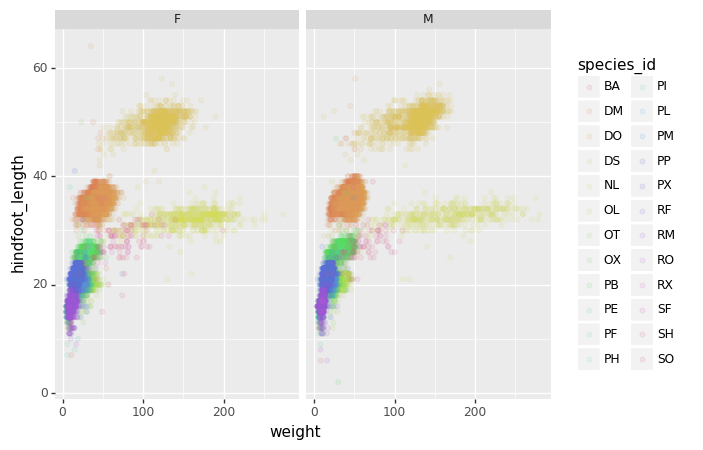
)We can now keep the same code and at the facet_wrap on a
chosen variable to split out the graph and make a separate graph for
each of the groups in that variable. As an example, use
sex:
PYTHON
(p9.ggplot(data=surveys_complete,
mapping=p9.aes(x='weight',
y='hindfoot_length',
color='species_id'))
+ p9.geom_point(alpha=0.1)
+ p9.facet_wrap("sex")
)
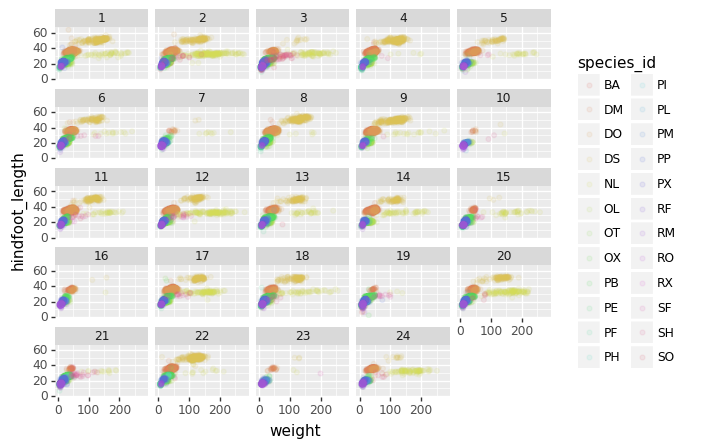
We can apply the same concept on any of the available categorical variables:
PYTHON
(p9.ggplot(data=surveys_complete,
mapping=p9.aes(x='weight',
y='hindfoot_length',
color='species_id'))
+ p9.geom_point(alpha=0.1)
+ p9.facet_wrap("plot_id")
)
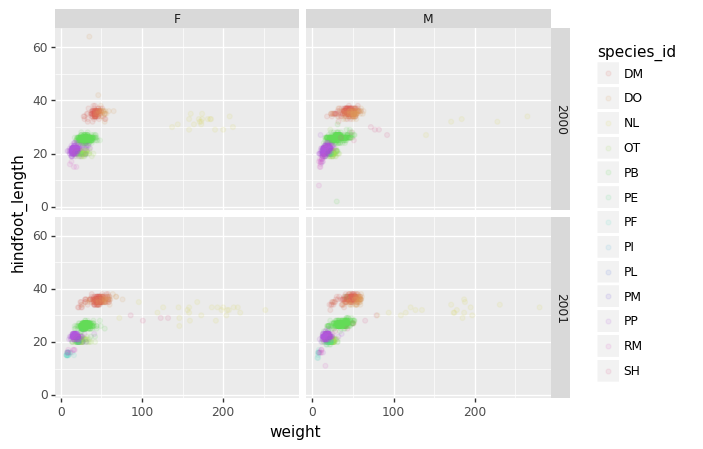
The facet_wrap geometry extracts plots into an arbitrary
number of dimensions to allow them to cleanly fit on one page. On the
other hand, the facet_grid geometry allows you to
explicitly specify how you want your plots to be arranged via formula
notation (rows ~ columns; a . can be used as a
placeholder that indicates only one row or column).
PYTHON
# only select the years of interest
survey_2000 = surveys_complete[surveys_complete["year"].isin([2000, 2001])]
(p9.ggplot(data=survey_2000,
mapping=p9.aes(x='weight',
y='hindfoot_length',
color='species_id'))
+ p9.geom_point(alpha=0.1)
+ p9.facet_grid("year ~ sex")
)
Challenge - facetting
Create a separate plot for each of the species that depicts how the average weight of the species changes through the years.
Challenge - facetting
Based on the previous exercise, visually compare how the weights of
male and females has changed through time by creating a separate plot
for each sex and an individual color assigned to each
species_id.
Further customization
As the syntax of plotnine follows the original R package
ggplot2, the documentation of ggplot2 can
provide information and inspiration to customize graphs. Take a look at
the ggplot2 cheat
sheet, and think of ways to improve the plot. You can write down
some of your ideas as comments in the Etherpad.
The theming options provide a rich set of visual adaptations. Consider the following example of a bar plot with the counts per year.
Notice that we use the year here as a categorical
variable by using the factor functionality. However, by
doing so, we have the individual year labels overlapping with each
other. The theme functionality provides a way to rotate the
text of the x-axis labels:
PYTHON
(p9.ggplot(data=surveys_complete,
mapping=p9.aes(x='factor(year)'))
+ p9.geom_bar()
+ p9.theme_bw()
+ p9.theme(axis_text_x = p9.element_text(angle=90))
)
When you like a specific set of theme-customizations you created, you can save them as an object to easily apply them to other plots you may create:
PYTHON
my_custom_theme = p9.theme(axis_text_x = p9.element_text(color="grey", size=10,
angle=90, hjust=.5),
axis_text_y = p9.element_text(color="grey", size=10))
(p9.ggplot(data=surveys_complete,
mapping=p9.aes(x='factor(year)'))
+ p9.geom_bar()
+ my_custom_theme
)
Challenge - customization
Please take another five minutes to either improve one of the plots generated in this exercise or create a beautiful graph of your own.
Here are some ideas:
- See if you can change thickness of lines for the line plot .
- Can you find a way to change the name of the legend? What about its labels?
- Use a different color palette (see http://www.cookbook-r.com/Graphs/Colors_(ggplot2))
After creating your plot, you can save it to a file in your favourite
format. You can easily change the dimension (and its resolution) of your
plot by adjusting the appropriate arguments (width,
height and dpi):
PYTHON
my_plot = (p9.ggplot(data=surveys_complete,
mapping=p9.aes(x='weight', y='hindfoot_length'))
+ p9.geom_point()
)
my_plot.save("scatterplot.png", width=10, height=10, dpi=300)Key Points
- The
data,aesvariables and ageometryare the main elements of a plotnine graph - With the
+operator, additionalscale_*,theme_*,xlab/ylabandfacet_*elements are added