Reproducibility
Last updated on 2023-02-13 | Edit this page
Estimated time: 10 minutes
Overview
Questions
- How to build reproducible analysis?
- how to deal with dependencies?
Objectives
- Describe the importance of version control systems
- Nudge the use of GitHub/GitLab for open collaboration
- Share open science practices for transparent and ethical research
Documentation as a guiding light for people who may feel lost otherwise. The Turing Way project illustration by Scriberia for The Turing Way Community Shared under CC-BY 4.0 License. Zenodo. http://doi.org/10.5281/zenodo.3332807
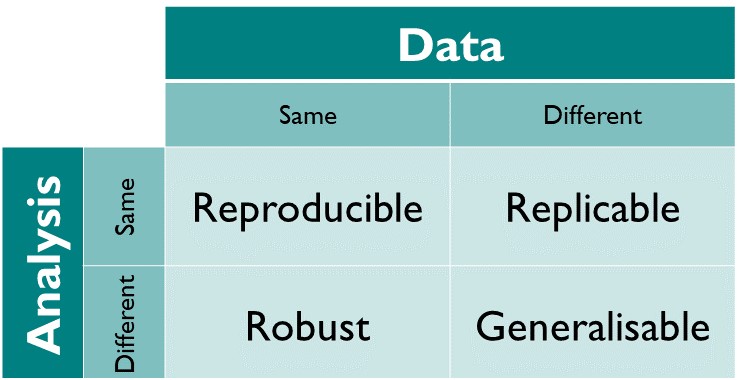
The different dimensions of reproducible research described in the matrix above have the following definitions directy taken from The Turing Way Guide to Reproducible Research (see the oveview chapter):
- Reproducible: A result is reproducible when the same analysis steps performed on the same dataset consistently produces the same answer.
- Replicable: A result is replicable when the same analysis performed on different datasets produces qualitatively similar answers.
- Robust: A result is robust when the same dataset is subjected to different analysis workflows to answer the same research question (for example one pipeline written in R and another written in Python) and a qualitatively similar or identical answer is produced. Robust results show that the work is not dependent on the specificities of the programming language chosen to perform the analysis.
- Generalisable: Combining replicable and robust findings allow us to form generalisable results. Note that running an analysis on a different software implementation and with a different dataset does not provide generalised results. There will be many more steps to know how well the work applies to all the different aspects of the research question. Generalisation is an important step towards understanding that the result is not dependent on a particular dataset nor a particular version of the analysis pipeline.
Thinking about which software, tools and platforms to use will greatly affect how you analyse and process data, as well as how you share your results for computational reproducibility. The idea is to facilitate others in recreating the setup process necessary to reproduce your research.
Some tools that can be used to enable these are the following:
- Dependency managers such as Conda keep dependencies updated and make sure the same version of dependencies used in the development environments are also used when reproducing a result.
- Containers such as Docker is a way to create computational environments with configurations required for developing, testing and using research software isolated/independent from other applications.
- Literate Programming using Jupyter Notebook is an extremely powerful way to use a web-based online interactive computing environment to execute code and script while adding notes and additional information about the application. To learn more about how to create a reproducible environment, the chapter on Reproducible Environments in The Turing way is a good place to start.
Reproducible Research Environment
Researchers’ working environments evolve as they update software, install new software, and move to different computers. If the project environment is not captured and the researchers need to return to their project after months or years (as is common in research), they will be unable to do so confidently. a computational environment is a system where a program is run. This includes features of hardware (such as the numbers of cores in any CPUs) and features of the software (such as the operating system, programming languages, supporting packages, other pieces of installed software, along with their versions and configurations).

There are several ways of capturing computational environments. The major ones covered in this chapter will be Package Management Systems, Binder, Virtual Machines, and Containers. Each has its pros and cons, and the most appropriate option for you will depend on the nature of your project. They can be broadly split into two categories: those that capture only the software and its versions used in an environment (Package Management Systems), and those that replicate an entire computational environment - including the operating system and customised settings (Virtual Machines and Containers).
Another way these can be split is by how the reproduced research is presented to the reproducer. Using Binder or a Virtual Machine creates a much more graphical, GUI-type result. In contrast, the outputs of Containers and Package Management Systems are more easily interacted with via the command line. Please read more about each of these concepts and their practice use, please visit Capturing Computational Environments in The Turing Way.
Key Points
- Version controlled repository help record different contributions and contributor information openly.
- Open Science is an umbrella term that involve different practices for research in the context of different research objects.
- Online Persistent Identifiers or Digital Object Identifiers are useful for releasing and citing different versions of research objects.
