Next steps
Last updated on 2023-07-11 | Edit this page
Estimated time 90 minutes
Overview
Questions
- SummarizedExperiment
Objectives
- Introduce the notion of data containers
- Give an overview of the
SummarizedExperiment, extensively used in omics analyses
Next steps
Data in bioinformatics is often complex. To deal with this, developers define specialised data containers (termed classes) that match the properties of the data they need to handle.
This aspect is central to the Bioconductor1 project which uses the same core data infrastructure across packages. This certainly contributed to Bioconductor’s success. Bioconductor package developers are advised to make use of existing infrastructure to provide coherence, interoperability, and stability to the project as a whole.
To illustrate such an omics data container, we’ll present the
SummarizedExperiment class.
SummarizedExperiment
The figure below represents the anatomy of the SummarizedExperiment class.
Objects of the class SummarizedExperiment contain :
One (or more) assay(s) containing the quantitative omics data (expression data), stored as a matrix-like object. Features (genes, transcripts, proteins, …) are defined along the rows, and samples along the columns.
A sample metadata slot containing sample co-variates, stored as a data frame. Rows from this table represent samples (rows match exactly the columns of the expression data).
A feature metadata slot containing feature co-variates, stored as a data frame. The rows of this data frame match exactly the rows of the expression data.
The coordinated nature of the SummarizedExperiment guarantees that during data manipulation, the dimensions of the different slots will always match (i.e the columns in the expression data and then rows in the sample metadata, as well as the rows in the expression data and feature metadata) during data manipulation. For example, if we had to exclude one sample from the assay, it would be automatically removed from the sample metadata in the same operation.
The metadata slots can grow additional co-variates (columns) without affecting the other structures.
Creating a SummarizedExperiment
In order to create a SummarizedExperiment, we will
create the individual components, i.e the count matrix, the sample and
gene metadata from csv files. These are typically how RNA-Seq data are
provided (after raw data have been processed).
-
An expression matrix: we load the count matrix,
specifying that the first columns contains row/gene names, and convert
the
data.frameto amatrix. You can download it here.
R
count_matrix <- read.csv("data/count_matrix.csv",
row.names = 1) %>%
as.matrix()
count_matrix[1:5, ]
OUTPUT
GSM2545336 GSM2545337 GSM2545338 GSM2545339 GSM2545340 GSM2545341
Asl 1170 361 400 586 626 988
Apod 36194 10347 9173 10620 13021 29594
Cyp2d22 4060 1616 1603 1901 2171 3349
Klk6 287 629 641 578 448 195
Fcrls 85 233 244 237 180 38
GSM2545342 GSM2545343 GSM2545344 GSM2545345 GSM2545346 GSM2545347
Asl 836 535 586 597 938 1035
Apod 24959 13668 13230 15868 27769 34301
Cyp2d22 3122 2008 2254 2277 2985 3452
Klk6 186 1101 537 567 327 233
Fcrls 68 375 199 177 89 67
GSM2545348 GSM2545349 GSM2545350 GSM2545351 GSM2545352 GSM2545353
Asl 494 481 666 937 803 541
Apod 11258 11812 15816 29242 20415 13682
Cyp2d22 1883 2014 2417 3678 2920 2216
Klk6 742 881 828 250 798 710
Fcrls 300 233 231 81 303 285
GSM2545354 GSM2545362 GSM2545363 GSM2545380
Asl 473 748 576 1192
Apod 11088 15916 11166 38148
Cyp2d22 1821 2842 2011 4019
Klk6 894 501 598 259
Fcrls 248 179 184 68R
dim(count_matrix)
OUTPUT
[1] 1474 22- A table describing the samples, available here.
R
sample_metadata <- read.csv("data/sample_metadata.csv")
sample_metadata
OUTPUT
sample organism age sex infection strain time tissue mouse
1 GSM2545336 Mus musculus 8 Female InfluenzaA C57BL/6 8 Cerebellum 14
2 GSM2545337 Mus musculus 8 Female NonInfected C57BL/6 0 Cerebellum 9
3 GSM2545338 Mus musculus 8 Female NonInfected C57BL/6 0 Cerebellum 10
4 GSM2545339 Mus musculus 8 Female InfluenzaA C57BL/6 4 Cerebellum 15
5 GSM2545340 Mus musculus 8 Male InfluenzaA C57BL/6 4 Cerebellum 18
6 GSM2545341 Mus musculus 8 Male InfluenzaA C57BL/6 8 Cerebellum 6
7 GSM2545342 Mus musculus 8 Female InfluenzaA C57BL/6 8 Cerebellum 5
8 GSM2545343 Mus musculus 8 Male NonInfected C57BL/6 0 Cerebellum 11
9 GSM2545344 Mus musculus 8 Female InfluenzaA C57BL/6 4 Cerebellum 22
10 GSM2545345 Mus musculus 8 Male InfluenzaA C57BL/6 4 Cerebellum 13
11 GSM2545346 Mus musculus 8 Male InfluenzaA C57BL/6 8 Cerebellum 23
12 GSM2545347 Mus musculus 8 Male InfluenzaA C57BL/6 8 Cerebellum 24
13 GSM2545348 Mus musculus 8 Female NonInfected C57BL/6 0 Cerebellum 8
14 GSM2545349 Mus musculus 8 Male NonInfected C57BL/6 0 Cerebellum 7
15 GSM2545350 Mus musculus 8 Male InfluenzaA C57BL/6 4 Cerebellum 1
16 GSM2545351 Mus musculus 8 Female InfluenzaA C57BL/6 8 Cerebellum 16
17 GSM2545352 Mus musculus 8 Female InfluenzaA C57BL/6 4 Cerebellum 21
18 GSM2545353 Mus musculus 8 Female NonInfected C57BL/6 0 Cerebellum 4
19 GSM2545354 Mus musculus 8 Male NonInfected C57BL/6 0 Cerebellum 2
20 GSM2545362 Mus musculus 8 Female InfluenzaA C57BL/6 4 Cerebellum 20
21 GSM2545363 Mus musculus 8 Male InfluenzaA C57BL/6 4 Cerebellum 12
22 GSM2545380 Mus musculus 8 Female InfluenzaA C57BL/6 8 Cerebellum 19R
dim(sample_metadata)
OUTPUT
[1] 22 9- A table describing the genes, available here.
R
gene_metadata <- read.csv("data/gene_metadata.csv")
gene_metadata[1:10, 1:4]
OUTPUT
gene ENTREZID
1 Asl 109900
2 Apod 11815
3 Cyp2d22 56448
4 Klk6 19144
5 Fcrls 80891
6 Slc2a4 20528
7 Exd2 97827
8 Gjc2 118454
9 Plp1 18823
10 Gnb4 14696
product
1 argininosuccinate lyase, transcript variant X1
2 apolipoprotein D, transcript variant 3
3 cytochrome P450, family 2, subfamily d, polypeptide 22, transcript variant 2
4 kallikrein related-peptidase 6, transcript variant 2
5 Fc receptor-like S, scavenger receptor, transcript variant X1
6 solute carrier family 2 (facilitated glucose transporter), member 4
7 exonuclease 3'-5' domain containing 2
8 gap junction protein, gamma 2, transcript variant 1
9 proteolipid protein (myelin) 1, transcript variant 1
10 guanine nucleotide binding protein (G protein), beta 4, transcript variant X2
ensembl_gene_id
1 ENSMUSG00000025533
2 ENSMUSG00000022548
3 ENSMUSG00000061740
4 ENSMUSG00000050063
5 ENSMUSG00000015852
6 ENSMUSG00000018566
7 ENSMUSG00000032705
8 ENSMUSG00000043448
9 ENSMUSG00000031425
10 ENSMUSG00000027669R
dim(gene_metadata)
OUTPUT
[1] 1474 9We will create a SummarizedExperiment from these
tables:
The count matrix that will be used as the
assayThe table describing the samples will be used as the sample metadata slot
The table describing the genes will be used as the features metadata slot
To do this we can put the different parts together using the
SummarizedExperiment constructor:
R
## BiocManager::install("SummarizedExperiment")
library("SummarizedExperiment")
WARNING
Warning: replacing previous import 'S4Arrays::makeNindexFromArrayViewport' by
'DelayedArray::makeNindexFromArrayViewport' when loading 'SummarizedExperiment'First, we make sure that the samples are in the same order in the count matrix and the sample annotation, and the same for the genes in the count matrix and the gene annotation.
R
stopifnot(rownames(count_matrix) == gene_metadata$gene)
stopifnot(colnames(count_matrix) == sample_metadata$sample)
R
se <- SummarizedExperiment(assays = list(counts = count_matrix),
colData = sample_metadata,
rowData = gene_metadata)
se
OUTPUT
class: SummarizedExperiment
dim: 1474 22
metadata(0):
assays(1): counts
rownames(1474): Asl Apod ... Lmx1a Pbx1
rowData names(9): gene ENTREZID ... phenotype_description
hsapiens_homolog_associated_gene_name
colnames(22): GSM2545336 GSM2545337 ... GSM2545363 GSM2545380
colData names(9): sample organism ... tissue mouseSaving data
Exporting data to a spreadsheet, as we did in a previous episode, has
several limitations, such as those described in the first chapter
(possible inconsistencies with , and . for
decimal separators and lack of variable type definitions). Furthermore,
exporting data to a spreadsheet is only relevant for rectangular data
such as dataframes and matrices.
A more general way to save data, that is specific to R and is
guaranteed to work on any operating system, is to use the
saveRDS function. Saving objects like this will generate a
binary representation on disk (using the rds file extension
here), which can be loaded back into R using the readRDS
function.
R
saveRDS(se, file = "data_output/se.rds")
rm(se)
se <- readRDS("data_output/se.rds")
head(se)
To conclude, when it comes to saving data from R that will be loaded
again in R, saving and loading with saveRDS and
readRDS is the preferred approach. If tabular data need to
be shared with somebody that is not using R, then exporting to a
text-based spreadsheet is a good alternative.
Using this data structure, we can access the expression matrix with
the assay function:
R
head(assay(se))
OUTPUT
GSM2545336 GSM2545337 GSM2545338 GSM2545339 GSM2545340 GSM2545341
Asl 1170 361 400 586 626 988
Apod 36194 10347 9173 10620 13021 29594
Cyp2d22 4060 1616 1603 1901 2171 3349
Klk6 287 629 641 578 448 195
Fcrls 85 233 244 237 180 38
Slc2a4 782 231 248 265 313 786
GSM2545342 GSM2545343 GSM2545344 GSM2545345 GSM2545346 GSM2545347
Asl 836 535 586 597 938 1035
Apod 24959 13668 13230 15868 27769 34301
Cyp2d22 3122 2008 2254 2277 2985 3452
Klk6 186 1101 537 567 327 233
Fcrls 68 375 199 177 89 67
Slc2a4 528 249 266 357 654 693
GSM2545348 GSM2545349 GSM2545350 GSM2545351 GSM2545352 GSM2545353
Asl 494 481 666 937 803 541
Apod 11258 11812 15816 29242 20415 13682
Cyp2d22 1883 2014 2417 3678 2920 2216
Klk6 742 881 828 250 798 710
Fcrls 300 233 231 81 303 285
Slc2a4 271 304 349 715 513 320
GSM2545354 GSM2545362 GSM2545363 GSM2545380
Asl 473 748 576 1192
Apod 11088 15916 11166 38148
Cyp2d22 1821 2842 2011 4019
Klk6 894 501 598 259
Fcrls 248 179 184 68
Slc2a4 248 350 317 796R
dim(assay(se))
OUTPUT
[1] 1474 22We can access the sample metadata using the colData
function:
R
colData(se)
OUTPUT
DataFrame with 22 rows and 9 columns
sample organism age sex infection
<character> <character> <integer> <character> <character>
GSM2545336 GSM2545336 Mus musculus 8 Female InfluenzaA
GSM2545337 GSM2545337 Mus musculus 8 Female NonInfected
GSM2545338 GSM2545338 Mus musculus 8 Female NonInfected
GSM2545339 GSM2545339 Mus musculus 8 Female InfluenzaA
GSM2545340 GSM2545340 Mus musculus 8 Male InfluenzaA
... ... ... ... ... ...
GSM2545353 GSM2545353 Mus musculus 8 Female NonInfected
GSM2545354 GSM2545354 Mus musculus 8 Male NonInfected
GSM2545362 GSM2545362 Mus musculus 8 Female InfluenzaA
GSM2545363 GSM2545363 Mus musculus 8 Male InfluenzaA
GSM2545380 GSM2545380 Mus musculus 8 Female InfluenzaA
strain time tissue mouse
<character> <integer> <character> <integer>
GSM2545336 C57BL/6 8 Cerebellum 14
GSM2545337 C57BL/6 0 Cerebellum 9
GSM2545338 C57BL/6 0 Cerebellum 10
GSM2545339 C57BL/6 4 Cerebellum 15
GSM2545340 C57BL/6 4 Cerebellum 18
... ... ... ... ...
GSM2545353 C57BL/6 0 Cerebellum 4
GSM2545354 C57BL/6 0 Cerebellum 2
GSM2545362 C57BL/6 4 Cerebellum 20
GSM2545363 C57BL/6 4 Cerebellum 12
GSM2545380 C57BL/6 8 Cerebellum 19R
dim(colData(se))
OUTPUT
[1] 22 9We can also access the feature metadata using the
rowData function:
R
head(rowData(se))
OUTPUT
DataFrame with 6 rows and 9 columns
gene ENTREZID product ensembl_gene_id
<character> <integer> <character> <character>
Asl Asl 109900 argininosuccinate ly.. ENSMUSG00000025533
Apod Apod 11815 apolipoprotein D, tr.. ENSMUSG00000022548
Cyp2d22 Cyp2d22 56448 cytochrome P450, fam.. ENSMUSG00000061740
Klk6 Klk6 19144 kallikrein related-p.. ENSMUSG00000050063
Fcrls Fcrls 80891 Fc receptor-like S, .. ENSMUSG00000015852
Slc2a4 Slc2a4 20528 solute carrier famil.. ENSMUSG00000018566
external_synonym chromosome_name gene_biotype phenotype_description
<character> <character> <character> <character>
Asl 2510006M18Rik 5 protein_coding abnormal circulating..
Apod NA 16 protein_coding abnormal lipid homeo..
Cyp2d22 2D22 15 protein_coding abnormal skin morpho..
Klk6 Bssp 7 protein_coding abnormal cytokine le..
Fcrls 2810439C17Rik 3 protein_coding decreased CD8-positi..
Slc2a4 Glut-4 11 protein_coding abnormal circulating..
hsapiens_homolog_associated_gene_name
<character>
Asl ASL
Apod APOD
Cyp2d22 CYP2D6
Klk6 KLK6
Fcrls FCRL2
Slc2a4 SLC2A4R
dim(rowData(se))
OUTPUT
[1] 1474 9Subsetting a SummarizedExperiment
SummarizedExperiment can be subset just like with data frames, with numerics or with characters of logicals.
Below, we create a new instance of class SummarizedExperiment that contains only the 5 first features for the 3 first samples.
R
se1 <- se[1:5, 1:3]
se1
OUTPUT
class: SummarizedExperiment
dim: 5 3
metadata(0):
assays(1): counts
rownames(5): Asl Apod Cyp2d22 Klk6 Fcrls
rowData names(9): gene ENTREZID ... phenotype_description
hsapiens_homolog_associated_gene_name
colnames(3): GSM2545336 GSM2545337 GSM2545338
colData names(9): sample organism ... tissue mouseR
colData(se1)
OUTPUT
DataFrame with 3 rows and 9 columns
sample organism age sex infection
<character> <character> <integer> <character> <character>
GSM2545336 GSM2545336 Mus musculus 8 Female InfluenzaA
GSM2545337 GSM2545337 Mus musculus 8 Female NonInfected
GSM2545338 GSM2545338 Mus musculus 8 Female NonInfected
strain time tissue mouse
<character> <integer> <character> <integer>
GSM2545336 C57BL/6 8 Cerebellum 14
GSM2545337 C57BL/6 0 Cerebellum 9
GSM2545338 C57BL/6 0 Cerebellum 10R
rowData(se1)
OUTPUT
DataFrame with 5 rows and 9 columns
gene ENTREZID product ensembl_gene_id
<character> <integer> <character> <character>
Asl Asl 109900 argininosuccinate ly.. ENSMUSG00000025533
Apod Apod 11815 apolipoprotein D, tr.. ENSMUSG00000022548
Cyp2d22 Cyp2d22 56448 cytochrome P450, fam.. ENSMUSG00000061740
Klk6 Klk6 19144 kallikrein related-p.. ENSMUSG00000050063
Fcrls Fcrls 80891 Fc receptor-like S, .. ENSMUSG00000015852
external_synonym chromosome_name gene_biotype phenotype_description
<character> <character> <character> <character>
Asl 2510006M18Rik 5 protein_coding abnormal circulating..
Apod NA 16 protein_coding abnormal lipid homeo..
Cyp2d22 2D22 15 protein_coding abnormal skin morpho..
Klk6 Bssp 7 protein_coding abnormal cytokine le..
Fcrls 2810439C17Rik 3 protein_coding decreased CD8-positi..
hsapiens_homolog_associated_gene_name
<character>
Asl ASL
Apod APOD
Cyp2d22 CYP2D6
Klk6 KLK6
Fcrls FCRL2We can also use the colData() function to subset on
something from the sample metadata or the rowData() to
subset on something from the feature metadata. For example, here we keep
only miRNAs and the non infected samples:
R
se1 <- se[rowData(se)$gene_biotype == "miRNA",
colData(se)$infection == "NonInfected"]
se1
OUTPUT
class: SummarizedExperiment
dim: 7 7
metadata(0):
assays(1): counts
rownames(7): Mir1901 Mir378a ... Mir128-1 Mir7682
rowData names(9): gene ENTREZID ... phenotype_description
hsapiens_homolog_associated_gene_name
colnames(7): GSM2545337 GSM2545338 ... GSM2545353 GSM2545354
colData names(9): sample organism ... tissue mouseR
assay(se1)
OUTPUT
GSM2545337 GSM2545338 GSM2545343 GSM2545348 GSM2545349 GSM2545353
Mir1901 45 44 74 55 68 33
Mir378a 11 7 9 4 12 4
Mir133b 4 6 5 4 6 7
Mir30c-2 10 6 16 12 8 17
Mir149 1 2 0 0 0 0
Mir128-1 4 1 2 2 1 2
Mir7682 2 0 4 1 3 5
GSM2545354
Mir1901 60
Mir378a 8
Mir133b 3
Mir30c-2 15
Mir149 2
Mir128-1 1
Mir7682 5R
colData(se1)
OUTPUT
DataFrame with 7 rows and 9 columns
sample organism age sex infection
<character> <character> <integer> <character> <character>
GSM2545337 GSM2545337 Mus musculus 8 Female NonInfected
GSM2545338 GSM2545338 Mus musculus 8 Female NonInfected
GSM2545343 GSM2545343 Mus musculus 8 Male NonInfected
GSM2545348 GSM2545348 Mus musculus 8 Female NonInfected
GSM2545349 GSM2545349 Mus musculus 8 Male NonInfected
GSM2545353 GSM2545353 Mus musculus 8 Female NonInfected
GSM2545354 GSM2545354 Mus musculus 8 Male NonInfected
strain time tissue mouse
<character> <integer> <character> <integer>
GSM2545337 C57BL/6 0 Cerebellum 9
GSM2545338 C57BL/6 0 Cerebellum 10
GSM2545343 C57BL/6 0 Cerebellum 11
GSM2545348 C57BL/6 0 Cerebellum 8
GSM2545349 C57BL/6 0 Cerebellum 7
GSM2545353 C57BL/6 0 Cerebellum 4
GSM2545354 C57BL/6 0 Cerebellum 2R
rowData(se1)
OUTPUT
DataFrame with 7 rows and 9 columns
gene ENTREZID product ensembl_gene_id
<character> <integer> <character> <character>
Mir1901 Mir1901 100316686 microRNA 1901 ENSMUSG00000084565
Mir378a Mir378a 723889 microRNA 378a ENSMUSG00000105200
Mir133b Mir133b 723817 microRNA 133b ENSMUSG00000065480
Mir30c-2 Mir30c-2 723964 microRNA 30c-2 ENSMUSG00000065567
Mir149 Mir149 387167 microRNA 149 ENSMUSG00000065470
Mir128-1 Mir128-1 387147 microRNA 128-1 ENSMUSG00000065520
Mir7682 Mir7682 102466847 microRNA 7682 ENSMUSG00000106406
external_synonym chromosome_name gene_biotype phenotype_description
<character> <character> <character> <character>
Mir1901 Mirn1901 18 miRNA NA
Mir378a Mirn378 18 miRNA abnormal mitochondri..
Mir133b mir 133b 1 miRNA no abnormal phenotyp..
Mir30c-2 mir 30c-2 1 miRNA NA
Mir149 Mirn149 1 miRNA increased circulatin..
Mir128-1 Mirn128 1 miRNA no abnormal phenotyp..
Mir7682 mmu-mir-7682 1 miRNA NA
hsapiens_homolog_associated_gene_name
<character>
Mir1901 NA
Mir378a MIR378A
Mir133b MIR133B
Mir30c-2 MIR30C2
Mir149 NA
Mir128-1 MIR128-1
Mir7682 NAR
assay(se)[1:3, colData(se)$time != 4]
OUTPUT
GSM2545336 GSM2545337 GSM2545338 GSM2545341 GSM2545342 GSM2545343
Asl 1170 361 400 988 836 535
Apod 36194 10347 9173 29594 24959 13668
Cyp2d22 4060 1616 1603 3349 3122 2008
GSM2545346 GSM2545347 GSM2545348 GSM2545349 GSM2545351 GSM2545353
Asl 938 1035 494 481 937 541
Apod 27769 34301 11258 11812 29242 13682
Cyp2d22 2985 3452 1883 2014 3678 2216
GSM2545354 GSM2545380
Asl 473 1192
Apod 11088 38148
Cyp2d22 1821 4019R
# Equivalent to
assay(se)[1:3, colData(se)$time == 0 | colData(se)$time == 8]
OUTPUT
GSM2545336 GSM2545337 GSM2545338 GSM2545341 GSM2545342 GSM2545343
Asl 1170 361 400 988 836 535
Apod 36194 10347 9173 29594 24959 13668
Cyp2d22 4060 1616 1603 3349 3122 2008
GSM2545346 GSM2545347 GSM2545348 GSM2545349 GSM2545351 GSM2545353
Asl 938 1035 494 481 937 541
Apod 27769 34301 11258 11812 29242 13682
Cyp2d22 2985 3452 1883 2014 3678 2216
GSM2545354 GSM2545380
Asl 473 1192
Apod 11088 38148
Cyp2d22 1821 4019R
rna |>
filter(gene %in% c("Asl", "Apod", "Cyd2d22")) |>
filter(time != 4) |> select(expression)
OUTPUT
# A tibble: 28 × 1
expression
<dbl>
1 1170
2 36194
3 361
4 10347
5 400
6 9173
7 988
8 29594
9 836
10 24959
# ℹ 18 more rowsThe long table and the SummarizedExperiment contain the
same information, but simply structured differently. Both have they
advantages: the former is a good fit for the tidyverse
packages, while the latter is the preferred structure for many
bioinformatics and statistical processing steps, such as a typical
RNA-Seq analyses using the DESeq2 package.
Adding variables to metadata
We can also add information to the metadata. Suppose that you want to add the center where the samples were collected…
R
colData(se)$center <- rep("University of Illinois", nrow(colData(se)))
colData(se)
OUTPUT
DataFrame with 22 rows and 10 columns
sample organism age sex infection
<character> <character> <integer> <character> <character>
GSM2545336 GSM2545336 Mus musculus 8 Female InfluenzaA
GSM2545337 GSM2545337 Mus musculus 8 Female NonInfected
GSM2545338 GSM2545338 Mus musculus 8 Female NonInfected
GSM2545339 GSM2545339 Mus musculus 8 Female InfluenzaA
GSM2545340 GSM2545340 Mus musculus 8 Male InfluenzaA
... ... ... ... ... ...
GSM2545353 GSM2545353 Mus musculus 8 Female NonInfected
GSM2545354 GSM2545354 Mus musculus 8 Male NonInfected
GSM2545362 GSM2545362 Mus musculus 8 Female InfluenzaA
GSM2545363 GSM2545363 Mus musculus 8 Male InfluenzaA
GSM2545380 GSM2545380 Mus musculus 8 Female InfluenzaA
strain time tissue mouse center
<character> <integer> <character> <integer> <character>
GSM2545336 C57BL/6 8 Cerebellum 14 University of Illinois
GSM2545337 C57BL/6 0 Cerebellum 9 University of Illinois
GSM2545338 C57BL/6 0 Cerebellum 10 University of Illinois
GSM2545339 C57BL/6 4 Cerebellum 15 University of Illinois
GSM2545340 C57BL/6 4 Cerebellum 18 University of Illinois
... ... ... ... ... ...
GSM2545353 C57BL/6 0 Cerebellum 4 University of Illinois
GSM2545354 C57BL/6 0 Cerebellum 2 University of Illinois
GSM2545362 C57BL/6 4 Cerebellum 20 University of Illinois
GSM2545363 C57BL/6 4 Cerebellum 12 University of Illinois
GSM2545380 C57BL/6 8 Cerebellum 19 University of IllinoisThis illustrates that the metadata slots can grow indefinitely without affecting the other structures!
tidySummarizedExperiment
You may be wondering, can we use tidyverse commands to interact with
SummarizedExperiment objects. The answer is yes, we can
with the tidySummarizedExperiment package.
Remember what our SummarizedExperiment object looks like.
R
se
OUTPUT
class: SummarizedExperiment
dim: 1474 22
metadata(0):
assays(1): counts
rownames(1474): Asl Apod ... Lmx1a Pbx1
rowData names(9): gene ENTREZID ... phenotype_description
hsapiens_homolog_associated_gene_name
colnames(22): GSM2545336 GSM2545337 ... GSM2545363 GSM2545380
colData names(10): sample organism ... mouse centerLoad tidySummarizedExperiment and then take a look at
the se object again.
R
#BiocManager::install("tidySummarizedExperiment")
library("tidySummarizedExperiment")
se
OUTPUT
# A SummarizedExperiment-tibble abstraction: 32,428 × 22
# [90mFeatures=1474 | Samples=22 | Assays=counts[0m
.feature .sample counts sample organism age sex infection strain time
<chr> <chr> <int> <chr> <chr> <int> <chr> <chr> <chr> <int>
1 Asl GSM2545336 1170 GSM25… Mus mus… 8 Fema… Influenz… C57BL… 8
2 Apod GSM2545336 36194 GSM25… Mus mus… 8 Fema… Influenz… C57BL… 8
3 Cyp2d22 GSM2545336 4060 GSM25… Mus mus… 8 Fema… Influenz… C57BL… 8
4 Klk6 GSM2545336 287 GSM25… Mus mus… 8 Fema… Influenz… C57BL… 8
5 Fcrls GSM2545336 85 GSM25… Mus mus… 8 Fema… Influenz… C57BL… 8
6 Slc2a4 GSM2545336 782 GSM25… Mus mus… 8 Fema… Influenz… C57BL… 8
7 Exd2 GSM2545336 1619 GSM25… Mus mus… 8 Fema… Influenz… C57BL… 8
8 Gjc2 GSM2545336 288 GSM25… Mus mus… 8 Fema… Influenz… C57BL… 8
9 Plp1 GSM2545336 43217 GSM25… Mus mus… 8 Fema… Influenz… C57BL… 8
10 Gnb4 GSM2545336 1071 GSM25… Mus mus… 8 Fema… Influenz… C57BL… 8
# ℹ 40 more rows
# ℹ 12 more variables: tissue <chr>, mouse <int>, center <chr>, gene <chr>,
# ENTREZID <int>, product <chr>, ensembl_gene_id <chr>,
# external_synonym <chr>, chromosome_name <chr>, gene_biotype <chr>,
# phenotype_description <chr>, hsapiens_homolog_associated_gene_name <chr>It’s still a SummarizedExperiment object, so maintains
the efficient structure, but now we can view it as a tibble. Note the
first line of the output says this, it’s a
SummarizedExperiment-tibble abstraction. We
can also see in the second line of the output the number of transcripts
and samples.
If we want to revert to the standard
SummarizedExperiment view we can do that.
R
options("restore_SummarizedExperiment_show" = TRUE)
se
OUTPUT
class: SummarizedExperiment
dim: 1474 22
metadata(0):
assays(1): counts
rownames(1474): Asl Apod ... Lmx1a Pbx1
rowData names(9): gene ENTREZID ... phenotype_description
hsapiens_homolog_associated_gene_name
colnames(22): GSM2545336 GSM2545337 ... GSM2545363 GSM2545380
colData names(10): sample organism ... mouse centerBut here we will use the tibble view.
R
options("restore_SummarizedExperiment_show" = FALSE)
se
OUTPUT
# A SummarizedExperiment-tibble abstraction: 32,428 × 22
# [90mFeatures=1474 | Samples=22 | Assays=counts[0m
.feature .sample counts sample organism age sex infection strain time
<chr> <chr> <int> <chr> <chr> <int> <chr> <chr> <chr> <int>
1 Asl GSM2545336 1170 GSM25… Mus mus… 8 Fema… Influenz… C57BL… 8
2 Apod GSM2545336 36194 GSM25… Mus mus… 8 Fema… Influenz… C57BL… 8
3 Cyp2d22 GSM2545336 4060 GSM25… Mus mus… 8 Fema… Influenz… C57BL… 8
4 Klk6 GSM2545336 287 GSM25… Mus mus… 8 Fema… Influenz… C57BL… 8
5 Fcrls GSM2545336 85 GSM25… Mus mus… 8 Fema… Influenz… C57BL… 8
6 Slc2a4 GSM2545336 782 GSM25… Mus mus… 8 Fema… Influenz… C57BL… 8
7 Exd2 GSM2545336 1619 GSM25… Mus mus… 8 Fema… Influenz… C57BL… 8
8 Gjc2 GSM2545336 288 GSM25… Mus mus… 8 Fema… Influenz… C57BL… 8
9 Plp1 GSM2545336 43217 GSM25… Mus mus… 8 Fema… Influenz… C57BL… 8
10 Gnb4 GSM2545336 1071 GSM25… Mus mus… 8 Fema… Influenz… C57BL… 8
# ℹ 40 more rows
# ℹ 12 more variables: tissue <chr>, mouse <int>, center <chr>, gene <chr>,
# ENTREZID <int>, product <chr>, ensembl_gene_id <chr>,
# external_synonym <chr>, chromosome_name <chr>, gene_biotype <chr>,
# phenotype_description <chr>, hsapiens_homolog_associated_gene_name <chr>We can now use tidyverse commands to interact with the SummarizedExperiment object.
We can use filter to filter for rows using a condition
e.g. to view all rows for one sample.
R
se %>% filter(.sample == "GSM2545336")
OUTPUT
# A SummarizedExperiment-tibble abstraction: 1,474 × 22
# [90mFeatures=1474 | Samples=1 | Assays=counts[0m
.feature .sample counts sample organism age sex infection strain time
<chr> <chr> <int> <chr> <chr> <int> <chr> <chr> <chr> <int>
1 Asl GSM2545336 1170 GSM25… Mus mus… 8 Fema… Influenz… C57BL… 8
2 Apod GSM2545336 36194 GSM25… Mus mus… 8 Fema… Influenz… C57BL… 8
3 Cyp2d22 GSM2545336 4060 GSM25… Mus mus… 8 Fema… Influenz… C57BL… 8
4 Klk6 GSM2545336 287 GSM25… Mus mus… 8 Fema… Influenz… C57BL… 8
5 Fcrls GSM2545336 85 GSM25… Mus mus… 8 Fema… Influenz… C57BL… 8
6 Slc2a4 GSM2545336 782 GSM25… Mus mus… 8 Fema… Influenz… C57BL… 8
7 Exd2 GSM2545336 1619 GSM25… Mus mus… 8 Fema… Influenz… C57BL… 8
8 Gjc2 GSM2545336 288 GSM25… Mus mus… 8 Fema… Influenz… C57BL… 8
9 Plp1 GSM2545336 43217 GSM25… Mus mus… 8 Fema… Influenz… C57BL… 8
10 Gnb4 GSM2545336 1071 GSM25… Mus mus… 8 Fema… Influenz… C57BL… 8
# ℹ 40 more rows
# ℹ 12 more variables: tissue <chr>, mouse <int>, center <chr>, gene <chr>,
# ENTREZID <int>, product <chr>, ensembl_gene_id <chr>,
# external_synonym <chr>, chromosome_name <chr>, gene_biotype <chr>,
# phenotype_description <chr>, hsapiens_homolog_associated_gene_name <chr>We can use select to specify columns we want to
view.
R
se %>% select(.sample)
OUTPUT
tidySummarizedExperiment says: Key columns are missing. A data frame is returned for independent data analysis.OUTPUT
# A tibble: 32,428 × 1
.sample
<chr>
1 GSM2545336
2 GSM2545336
3 GSM2545336
4 GSM2545336
5 GSM2545336
6 GSM2545336
7 GSM2545336
8 GSM2545336
9 GSM2545336
10 GSM2545336
# ℹ 32,418 more rowsWe can use mutate to add metadata info.
R
se %>% mutate(center = "Heidelberg University")
OUTPUT
# A SummarizedExperiment-tibble abstraction: 32,428 × 22
# [90mFeatures=1474 | Samples=22 | Assays=counts[0m
.feature .sample counts sample organism age sex infection strain time
<chr> <chr> <int> <chr> <chr> <int> <chr> <chr> <chr> <int>
1 Asl GSM2545336 1170 GSM25… Mus mus… 8 Fema… Influenz… C57BL… 8
2 Apod GSM2545336 36194 GSM25… Mus mus… 8 Fema… Influenz… C57BL… 8
3 Cyp2d22 GSM2545336 4060 GSM25… Mus mus… 8 Fema… Influenz… C57BL… 8
4 Klk6 GSM2545336 287 GSM25… Mus mus… 8 Fema… Influenz… C57BL… 8
5 Fcrls GSM2545336 85 GSM25… Mus mus… 8 Fema… Influenz… C57BL… 8
6 Slc2a4 GSM2545336 782 GSM25… Mus mus… 8 Fema… Influenz… C57BL… 8
7 Exd2 GSM2545336 1619 GSM25… Mus mus… 8 Fema… Influenz… C57BL… 8
8 Gjc2 GSM2545336 288 GSM25… Mus mus… 8 Fema… Influenz… C57BL… 8
9 Plp1 GSM2545336 43217 GSM25… Mus mus… 8 Fema… Influenz… C57BL… 8
10 Gnb4 GSM2545336 1071 GSM25… Mus mus… 8 Fema… Influenz… C57BL… 8
# ℹ 40 more rows
# ℹ 12 more variables: tissue <chr>, mouse <int>, center <chr>, gene <chr>,
# ENTREZID <int>, product <chr>, ensembl_gene_id <chr>,
# external_synonym <chr>, chromosome_name <chr>, gene_biotype <chr>,
# phenotype_description <chr>, hsapiens_homolog_associated_gene_name <chr>We can also combine commands with the tidyverse pipe
%>%. For example, we could combine group_by
and summarise to get the total counts for each sample.
R
se %>%
group_by(.sample) %>%
summarise(total_counts=sum(counts))
OUTPUT
tidySummarizedExperiment says: A data frame is returned for independent data analysis.OUTPUT
# A tibble: 22 × 2
.sample total_counts
<chr> <int>
1 GSM2545336 3039671
2 GSM2545337 2602360
3 GSM2545338 2458618
4 GSM2545339 2500082
5 GSM2545340 2479024
6 GSM2545341 2413723
7 GSM2545342 2349728
8 GSM2545343 3105652
9 GSM2545344 2524137
10 GSM2545345 2506038
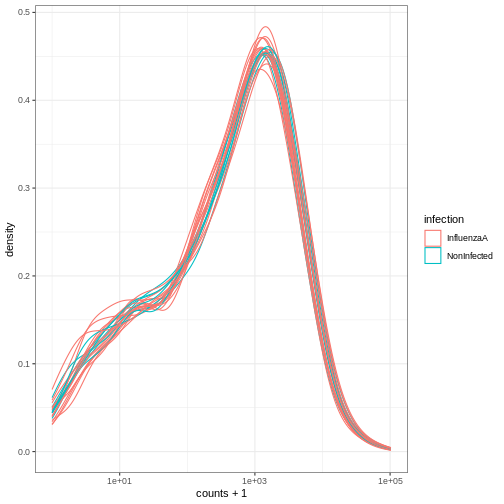
# ℹ 12 more rowsWe can treat the tidy SummarizedExperiment object as a normal tibble for plotting.
Here we plot the distribution of counts per sample.
R
se %>%
ggplot(aes(counts + 1, group=.sample, color=infection)) +
geom_density() +
scale_x_log10() +
theme_bw()
For more information on tidySummarizedExperiment, see the package website here.
Take-home message
SummarizedExperimentrepresents an efficient way to store and handle omics data.They are used in many Bioconductor packages.
If you follow the next training focused on RNA sequencing analysis,
you will learn to use the Bioconductor DESeq2 package to do
some differential expression analyses. The whole analysis of the
DESeq2 package is handled in a
SummarizedExperiment.
The Bioconductor was initiated by Robert Gentleman, one of the two creators of the R language. Bioconductor provides tools dedicated to omics data analysis. Bioconductor uses the R statistical programming language and is open source and open development.↩︎
